Dr. Crispy the Potato
Prerequisites
This is a practical project that assumes you have already covered the following concepts:
Visualize Dataset
for dir in "$(pwd)"/*; do
label=$(basename "$dir")
count=$(find "$dir" -type f -name "*.JPG" | wc -l)
echo "\"$label\": $count"
done
"Potato___Early_blight": 1000
"Potato___Late_blight": 1000
"Potato___healthy": 152
Early Blight








Late Blight








Healthy








Visualize Wrong predictions
| Late Blight predected Healthy | Late Blight predected Healthy | Late Blight predected Early Blight |
|---|---|---|
 |  |  |
| Late Blight predected Healthy | Late Blight predected Healthy |
|---|---|
 |  |
Output
Confusion Matrix
Accuracy/Loss Graph
Logs
[2023-10-19 21:16:49] Train size: 54, Validation size: 6, Test size: 8
[2023-10-19 21:16:49] Training model for 50 epochs.
[2023-10-19 21:48:43] Training took 1,914.36 seconds.
[2023-10-19 21:53:24] Loss: 0.0487, Accuracy: 98.05%
[2023-10-19 22:32:04] Number of wrong predictions: 5 / 256
[2023-10-19 22:32:04] Accuracy: 98.05%
[2023-10-26 11:12:11] Train size: 54, Validation size: 6, Test size: 8
Important Concepts
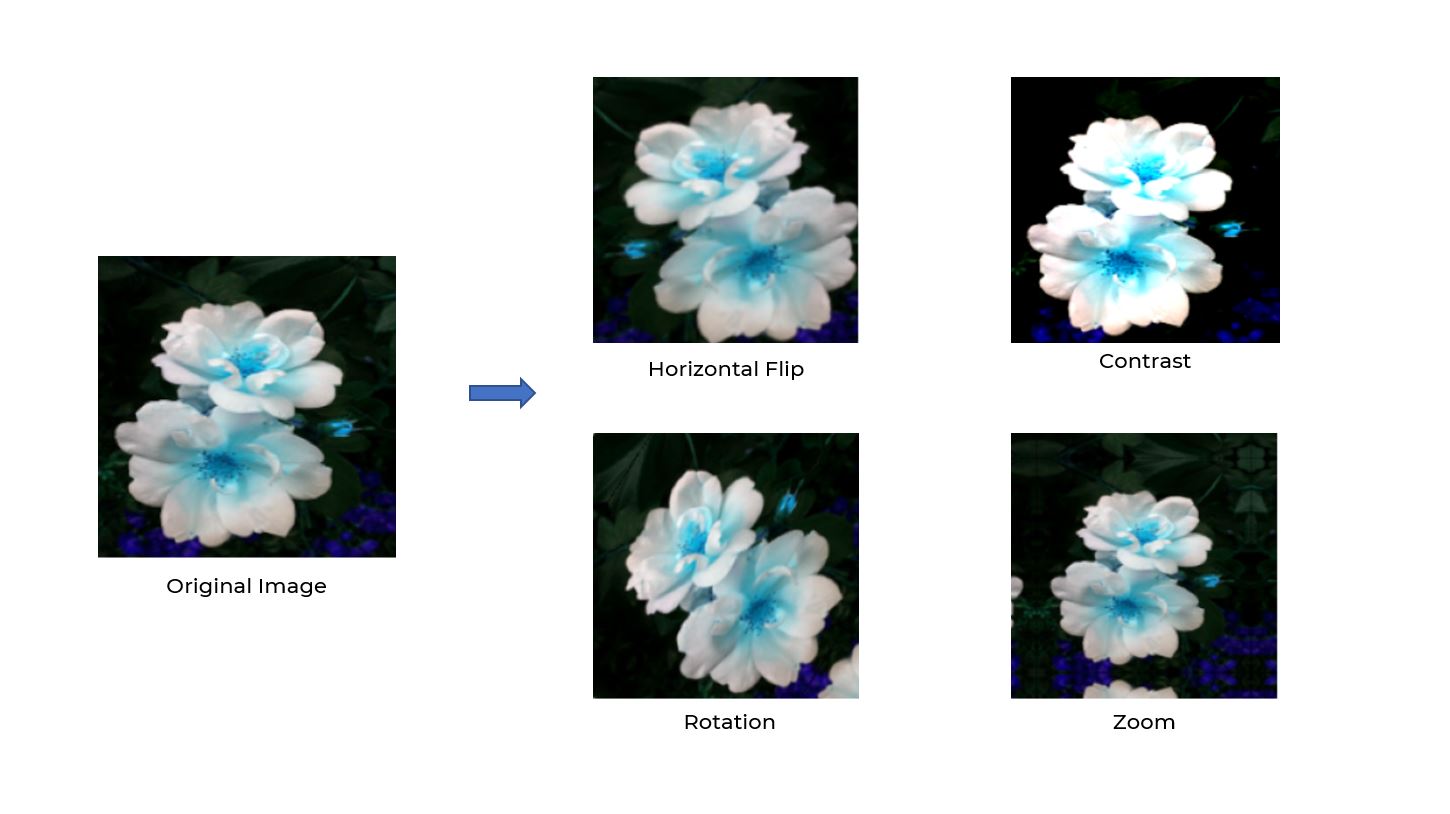
Data Augmentation
- Data augmentation is a technique to artificially create new training data from existing training data.
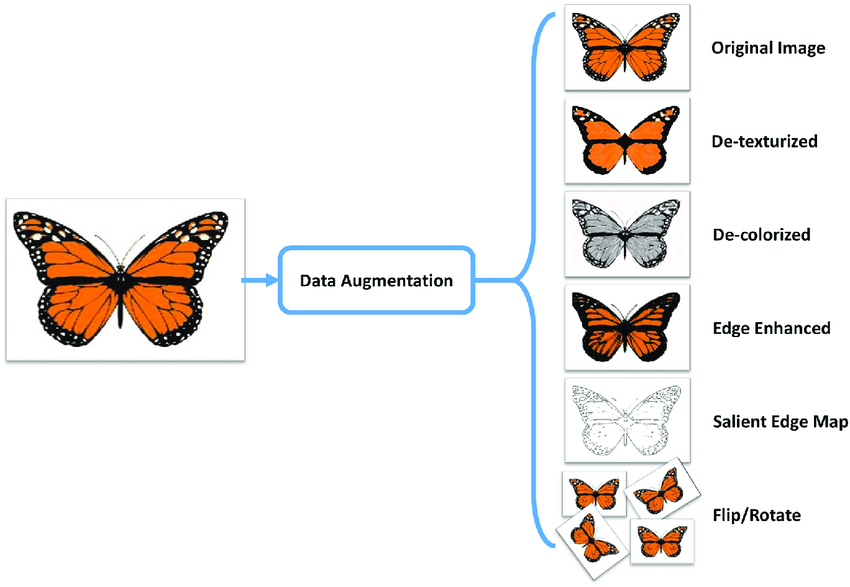
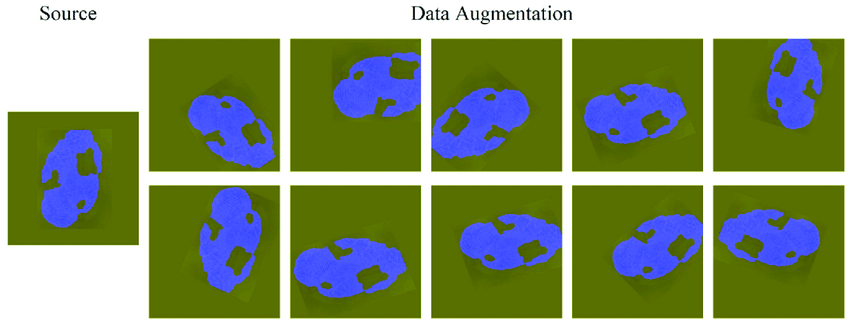
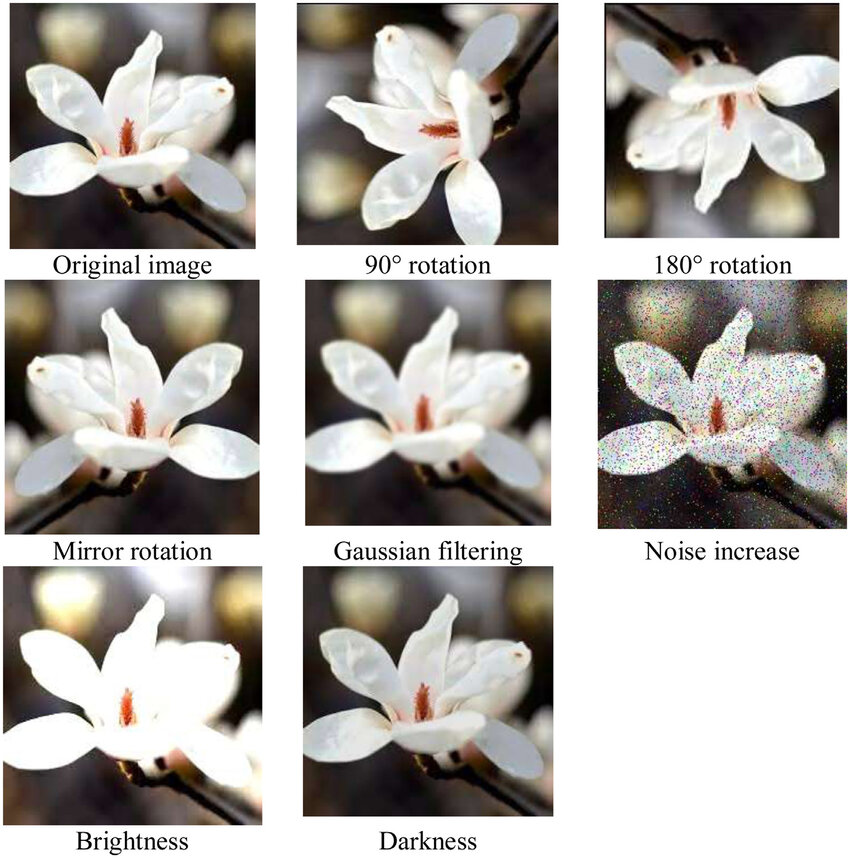
Code
Import and Magic Numbers
import os
from typing import Final
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
import utils
EPOCHS: Final[int] = 50
utils.py
BASE_DIR: Final[Path] = Path(__file__).resolve().parent.parent.parent
DATA_DIR: Final[Path] = os.path.join(BASE_DIR, "data")
LOGGING_DIR: Final[Path] = os.path.join(BASE_DIR, "logs")
TEST_DIR: Final[Path] = os.path.join(DATA_DIR, "test")
IMG_HEIGHT: Final[int] = 256
IMG_WIDTH: Final[int] = 256
CHANNELS: Final[int] = 3
BATCH_SIZE: Final[int] = 32
def show_image(image: np.ndarray, title: str) -> None:
cv2.startWindowThread()
cv2.imshow(title, image)
while cv2.waitKey(1) & 0xFF != ord("q"):
pass
cv2.destroyWindow(title)
# cv2 on Mac M1 is still buggy, so we need to wait a bit
for _ in range(5):
cv2.waitKey(1)
def log_to_file(message: str, custom_file: str="tune.log") -> None:
timestamp = datetime.datetime.now().strftime("%Y-%m-%d %H:%M:%S")
log_message = f"[{timestamp}] {message}"
logs_file_path: Path = os.path.join(LOGGING_DIR, custom_file)
with open(logs_file_path, "a") as f:
f.write(f"{log_message}\n")
Load Dataset
dataset: tf.data.Dataset = tf.keras.preprocessing.image_dataset_from_directory(
os.path.join(utils.DATA_DIR, 'dataset'),
shuffle=True,
image_size=(utils.IMG_HEIGHT, utils.IMG_WIDTH),
batch_size=utils.BATCH_SIZE
)
Found 2152 files belonging to 3 classes.
class_names: list[str] = dataset.class_names
print(class_names)
['Potato___Early_blight', 'Potato___Late_blight', 'Potato___healthy']
Preprocessing
- 80% "1721 Images or 54 Batches" of the data is used for training
- 10% "0215 Images or 06 Batches" of the data is used for validation
- 10% "0216 Images or 08 Batches" of the data is used for testing
-- total: 2152 images
def get_dataset_partitions(
dataset: tf.data.Dataset,
train_split: float=0.8,
validation_split: float=0.1,
test_split: float=0.1,
shuffle: bool=True
) -> tuple[tf.data.Dataset, tf.data.Dataset, tf.data.Dataset]:
"""Split dataset into train, validation and test partitions."""
DATASET_SIZE: Final[int] = dataset.cardinality().numpy()
if shuffle:
import random
dataset = dataset.shuffle(
buffer_size=dataset.cardinality(),
seed=random.randint(0, 10_000)
)
train_size = int(train_split * DATASET_SIZE)
val_size = int(validation_split * DATASET_SIZE)
train_dataset = dataset.take(train_size)
validation_dataset = dataset.skip(train_size).take(val_size)
test_dataset = dataset.skip(train_size + val_size)
utils.log_to_file(
f"Train size: {len(train_dataset)}, Validation size: {len(validation_dataset)}, Test size: {len(test_dataset)}"
)
return train_dataset, validation_dataset, test_dataset
train_dataset, validation_dataset, test_dataset = get_dataset_partitions(dataset)
Shuffle and Cache
train_dataset = train_dataset.cache().shuffle(
buffer_size=train_dataset.cardinality()
).prefetch(
buffer_size=tf.data.AUTOTUNE
)
validation_dataset = validation_dataset.cache().shuffle(
buffer_size=validation_dataset.cardinality()
).prefetch(
buffer_size=tf.data.AUTOTUNE
)
test_dataset = test_dataset.cache().shuffle(
buffer_size=test_dataset.cardinality()
).prefetch(
buffer_size=tf.data.AUTOTUNE
)
Normalize and Data Augmentation
# Normalize the data
resize_rescale = tf.keras.Sequential([
tf.keras.layers.experimental.preprocessing.Resizing(
utils.IMG_HEIGHT,
utils.IMG_WIDTH
),
tf.keras.layers.experimental.preprocessing.Rescaling(1.0/255)
])
# Data augmentation
data_augmentation = tf.keras.Sequential([
tf.keras.layers.experimental.preprocessing.RandomFlip(
"horizontal_and_vertical"
),
tf.keras.layers.experimental.preprocessing.RandomRotation(0.2)
])
Training
input_shape = (
utils.BATCH_SIZE,
utils.IMG_HEIGHT,
utils.IMG_WIDTH,
utils.CHANNELS
)
model = tf.keras.Sequential([
resize_rescale,
data_augmentation,
tf.keras.layers.Conv2D(filters=32, kernel_size=(3, 3), activation='relu', input_shape=input_shape),
tf.keras.layers.MaxPooling2D(pool_size=(2, 2)),
tf.keras.layers.Conv2D(filters=64, kernel_size=(3, 3), activation='relu'),
tf.keras.layers.MaxPooling2D(pool_size=(2, 2)),
tf.keras.layers.Conv2D(filters=64, kernel_size=(3, 3), activation='relu'),
tf.keras.layers.MaxPooling2D(pool_size=(2, 2)),
tf.keras.layers.Conv2D(filters=64, kernel_size=(3, 3), activation='relu'),
tf.keras.layers.MaxPooling2D(pool_size=(2, 2)),
tf.keras.layers.Conv2D(filters=64, kernel_size=(3, 3), activation='relu'),
tf.keras.layers.MaxPooling2D(pool_size=(2, 2)),
tf.keras.layers.Conv2D(filters=64, kernel_size=(3, 3), activation='relu'),
tf.keras.layers.MaxPooling2D(pool_size=(2, 2)),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(units=64, activation='relu'),
tf.keras.layers.Dense(units=3, activation='softmax')
])
model.build(input_shape=input_shape)
model.compile(
optimizer=tf.keras.optimizers.Adam(),
loss=tf.keras.losses.SparseCategoricalCrossentropy(),
metrics=['accuracy']
)
utils.log_to_file(f"Training model for {EPOCHS} epochs.")
import time
start_time = time.time()
history = model.fit(
train_dataset,
epochs=EPOCHS,
validation_data=validation_dataset
)
utils.log_to_file(f"Training took {(time.time() - start_time):,.2f} seconds.")
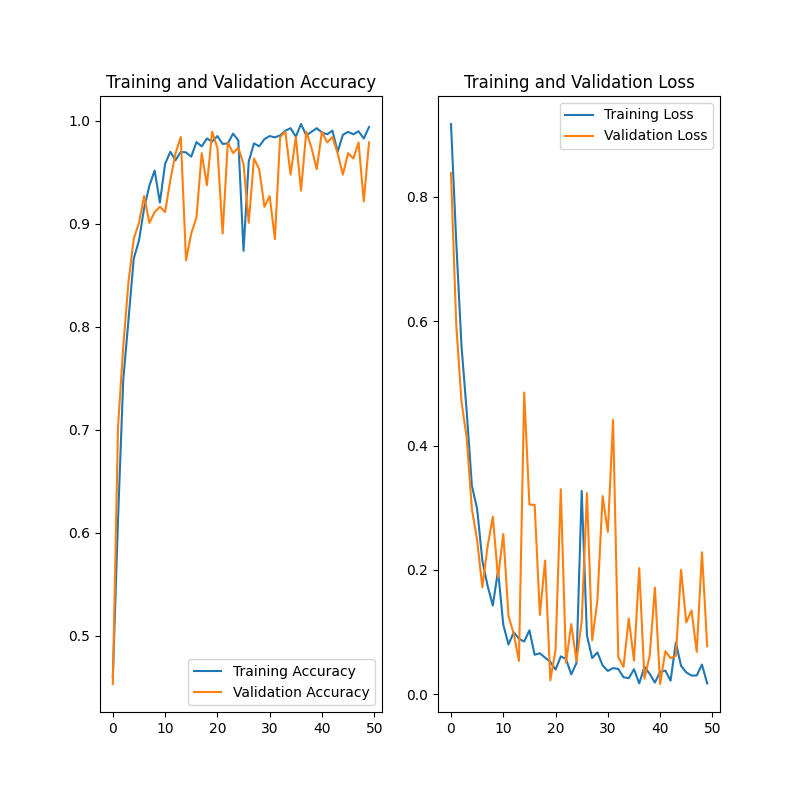
accuracy_history = history.history['accuracy']
val_accuracy_history = history.history['val_accuracy']
loss_history = history.history['loss']
val_loss_history = history.history['val_loss']
plt.figure(figsize=(8, 8))
plt.subplot(1, 2, 1)
plt.plot(range(EPOCHS), accuracy_history, label='Training Accuracy')
plt.plot(range(EPOCHS), val_accuracy_history, label='Validation Accuracy')
plt.legend(loc='lower right')
plt.title('Training and Validation Accuracy')
plt.subplot(1, 2, 2)
plt.plot(range(EPOCHS), loss_history, label='Training Loss')
plt.plot(range(EPOCHS), val_loss_history, label='Validation Loss')
plt.legend(loc='upper right')
plt.title('Training and Validation Loss')
plt.savefig(os.path.join(utils.DATA_DIR, "output", "accuracy.png"))

loss, accuracy = model.evaluate(test_dataset)
utils.log_to_file(f"Loss: {loss:,.4f}, Accuracy: {accuracy*100:,.2f}%")
model.summary()
Model: "sequential_2"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
sequential (Sequential) (None, 256, 256, 3) 0
sequential_1 (Sequential) (None, 256, 256, 3) 0
conv2d (Conv2D) (None, 254, 254, 32) 896
max_pooling2d (MaxPooling2 (None, 127, 127, 32) 0
D)
conv2d_1 (Conv2D) (None, 125, 125, 64) 18496
max_pooling2d_1 (MaxPoolin (None, 62, 62, 64) 0
g2D)
conv2d_2 (Conv2D) (None, 60, 60, 64) 36928
max_pooling2d_2 (MaxPoolin (None, 30, 30, 64) 0
g2D)
conv2d_3 (Conv2D) (None, 28, 28, 64) 36928
max_pooling2d_3 (MaxPoolin (None, 14, 14, 64) 0
g2D)
conv2d_4 (Conv2D) (None, 12, 12, 64) 36928
max_pooling2d_4 (MaxPoolin (None, 6, 6, 64) 0
g2D)
conv2d_5 (Conv2D) (None, 4, 4, 64) 36928
max_pooling2d_5 (MaxPoolin (None, 2, 2, 64) 0
g2D)
flatten (Flatten) (None, 256) 0
dense (Dense) (None, 64) 16448
dense_1 (Dense) (None, 3) 195
=================================================================
Total params: 183747 (717.76 KB)
Trainable params: 183747 (717.76 KB)
Non-trainable params: 0 (0.00 Byte)
_________________________________________________________________
model.save(os.path.join(
utils.DATA_DIR,
'models',
f"potato_model_{EPOCHS}_{accuracy*100:.2f}.model"
))
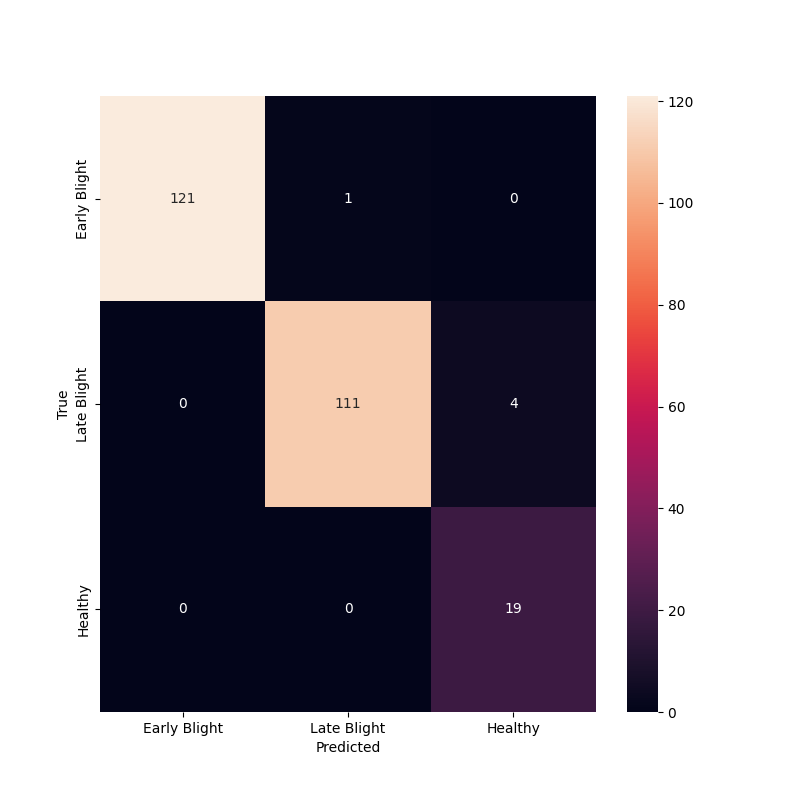
Confusion Matrix
from sklearn.metrics import confusion_matrix
import seaborn as sns
y_pred = []
y_true = []
for images_batch, label_batch in test_dataset:
for image, label in zip(images_batch, label_batch):
y_pred.append(np.argmax(model.predict(image[np.newaxis, ...])))
y_true.append(label.numpy())
cm = confusion_matrix(y_true, y_pred)
utils.log_to_file(
f"Number of wrong predictions: {(len(y_true) - np.trace(cm)):,} / {len(y_true):,}"
)
utils.log_to_file(f"Accuracy: {(np.trace(cm) / len(y_true))*100:.2f}%")
class_names = ['Early Blight', 'Late Blight', 'Healthy']
plt.figure(figsize=(8, 8))
sns.heatmap(cm, annot=True, fmt='g', xticklabels=class_names, yticklabels=class_names)
plt.xlabel("Predicted")
plt.ylabel("True")
plt.savefig(os.path.join(
utils.DATA_DIR,
"output",
"confusion_matrix.png"
))

Wrong predictions
wrong_predictions = []
for images_batch, label_batch in test_dataset:
wrong_predictions.extend(
(image.numpy().astype("uint8"), label.numpy(), np.argmax(model.predict(image[np.newaxis, ...])))
for image, label in zip(images_batch, label_batch)
if np.argmax(model.predict(image[np.newaxis, ...])) != label.numpy()
)
print(len(wrong_predictions))
5
for i, (image, true_label, predicted_label) in enumerate(wrong_predictions):
image_path: Path = os.path.join(
utils.BASE_DIR,
"temp",
"wrong-predictions",
f"{i:02d}-{class_names[true_label]}-{class_names[predicted_label]}.png"
)
os.makedirs(os.path.dirname(image_path), exist_ok=True)
cv2.imwrite(str(image_path), image)
| Late Blight predected Healthy | Late Blight predected Healthy | Late Blight predected Early Blight |
|---|---|---|
 |  |  |
| Late Blight predected Healthy | Late Blight predected Healthy |
|---|---|
 |  |
Logs
[2023-10-19 21:16:49] Train size: 54, Validation size: 6, Test size: 8
[2023-10-19 21:16:49] Training model for 50 epochs.
[2023-10-19 21:48:43] Training took 1,914.36 seconds.
[2023-10-19 21:53:24] Loss: 0.0487, Accuracy: 98.05%
[2023-10-19 22:32:04] Number of wrong predictions: 5 / 256
[2023-10-19 22:32:04] Accuracy: 98.05%
[2023-10-26 11:12:11] Train size: 54, Validation size: 6, Test size: 8


